Biology:DNA glycosylase
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first step of this process. They remove the damaged nitrogenous base while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site. This is accomplished by flipping the damaged base out of the double helix followed by cleavage of the N-glycosidic bond.[1]
Glycosylases were first discovered in bacteria, and have since been found in all kingdoms of life. In addition to their role in base excision repair, DNA glycosylase enzymes have been implicated in the repression of gene silencing in A. thaliana, N. tabacum and other plants by active demethylation. 5-methylcytosine residues are excised and replaced with unmethylated cytosines allowing access to the chromatin structure of the enzymes and proteins necessary for transcription and subsequent translation.[2][3]
Monofunctional vs. bifunctional glycosylases
There are two main classes of glycosylases: monofunctional and bifunctional. Monofunctional glycosylases have only glycosylase activity, whereas bifunctional glycosylases also possess AP lyase activity that permits them to cut the phosphodiester bond of DNA, creating a single-strand break without the need for an AP endonuclease. β-Elimination of an AP site by a glycosylase-lyase yields a 3' α,β-unsaturated aldehyde adjacent to a 5' phosphate, which differs from the AP endonuclease cleavage product.[4] Some glycosylase-lyases can further perform δ-elimination, which converts the 3' aldehyde to a 3' phosphate.
Biochemical mechanism
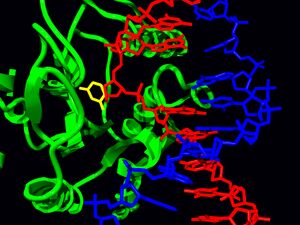
The first crystal structure of a DNA glycosylase was obtained for E. coli Nth.[5] This structure revealed that the enzyme flips the damaged base out of the double helix into an active site pocket in order to excise it. Other glycosylases have since been found to follow the same general paradigm, including human UNG pictured below. To cleave the N-glycosidic bond, monofunctional glycosylases use an activated water molecule to attack carbon 1 of the substrate. Bifunctional glycosylases, instead, use an amine residue as a nucleophile to attack the same carbon, going through a Schiff base intermediate.
Types of glycosylases
Crystal structures of many glycosylases have been solved. Based on structural similarity, glycosylases are grouped into four superfamilies. The UDG and AAG families contain small, compact glycosylases, whereas the MutM/Fpg and HhH-GPD families comprise larger enzymes with multiple domains.[4]
A wide variety of glycosylases have evolved to recognize different damaged bases. The table below summarizes the properties of known glycosylases in commonly studied model organisms.
| E. coli | B. cereus | Yeast (S. cerevisiae) | Human | Type | Substrates |
|---|---|---|---|---|---|
| AlkA | AlkE | Mag1 | MPG (N-methylpurine DNA glycosylase) | monofunctional | 3-meA(3-alkyladenine), hypoxanthine |
| UDG | Ung1 | UNG | monofunctional | uracil | |
| Fpg | Ogg1 | hOGG1 | bifunctional | 8-oxoG (8-Oxoguanine), FapyG | |
| Nth | Ntg1 | hNTH1 | bifunctional | Tg, hoU, hoC, urea, FapyG(2,6-diamino-4-hydroxy-5-formamidopyrimidine) | |
| Ntg2 | |||||
| Nei | Not present | hNEIL1 | bifunctional | Tg, hoU, hoC, urea, FapyG, FapyA(4,6-diamino-5-formamidopyrimidine) | |
| hNEIL2 | AP site, hoU | ||||
| hNEIL3 | unknown | ||||
| MutY | Not present | hMYH | monofunctional | A:8-oxoG | |
| Not present | Not present | hSMUG1 | monofunctional | U, hoU(5-hydroxyuracil), hmU(5-hydroxymethyluracil), fU(5-formyluracil) | |
| Not present | Not present | TDG | monofunctional | T:G mispair | |
| Not present | Not present | MBD4 | monofunctional | T:G mispair | |
| AlkC | AlkC | Not present | Not present | monofunctional | Alkylpurine |
| AlkD | AlkD | Not present | Not present | monofunctional | Alkylpurine |
DNA glycosylases can be grouped into the following categories based on their substrate(s):
Uracil DNA glycosylases
In molecular biology, the protein family, Uracil-DNA glycosylase (UDG) is an enzyme that reverts mutations in DNA. The most common mutation is the deamination of cytosine to uracil. UDG repairs these mutations. UDG is crucial in DNA repair, without it these mutations may lead to cancer.[8]
This entry represents various uracil-DNA glycosylases and related DNA glycosylases (EC), such as uracil-DNA glycosylase,[9] thermophilic uracil-DNA glycosylase,[10] G:T/U mismatch-specific DNA glycosylase (Mug),[11] and single-strand selective monofunctional uracil-DNA glycosylase (SMUG1).[12]
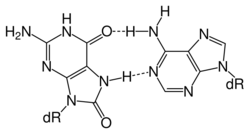
Uracil DNA glycosylases remove uracil from DNA, which can arise either by spontaneous deamination of cytosine or by the misincorporation of dU opposite dA during DNA replication. The prototypical member of this family is E. coli UDG, which was among the first glycosylases discovered. Four different uracil-DNA glycosylase activities have been identified in mammalian cells, including UNG, SMUG1, TDG, and MBD4. They vary in substrate specificity and subcellular localization. SMUG1 prefers single-stranded DNA as substrate, but also removes U from double-stranded DNA. In addition to unmodified uracil, SMUG1 can excise 5-hydroxyuracil, 5-hydroxymethyluracil and 5-formyluracil bearing an oxidized group at ring C5.[13] TDG and MBD4 are strictly specific for double-stranded DNA. TDG can remove thymine glycol when present opposite guanine, as well as derivatives of U with modifications at carbon 5. Current evidence suggests that, in human cells, TDG and SMUG1 are the major enzymes responsible for the repair of the U:G mispairs caused by spontaneous cytosine deamination, whereas uracil arising in DNA through dU misincorporation is mainly dealt with by UNG. MBD4 is thought to correct T:G mismatches that arise from deamination of 5-methylcytosine to thymine in CpG sites.[14] MBD4 mutant mice develop normally and do not show increased cancer susceptibility or reduced survival. But they acquire more C T mutations at CpG sequences in epithelial cells of the small intestine.[15]
The structure of human UNG in complex with DNA revealed that, like other glycosylases, it flips the target nucleotide out of the double helix and into the active site pocket.[16] UDG undergoes a conformational change from an ‘‘open’’ unbound state to a ‘‘closed’’ DNA-bound state.[17]
| UDG | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Epstein–Barr virus uracil-dna glycosylase in complex with ugi from pbs-2 | |||||||||
| Identifiers | |||||||||
| Symbol | UDG | ||||||||
| Pfam | PF03167 | ||||||||
| InterPro | IPR005122 | ||||||||
| PROSITE | PDOC00121 | ||||||||
| SCOP2 | 1udg / SCOPe / SUPFAM | ||||||||
| CDD | cd09593 | ||||||||
| |||||||||
History
Lindahl was the first to observe repair of uracil in DNA. UDG was purified from Escherichia coli, and this hydrolysed the N-glycosidic bond connecting the base to the deoxyribose sugar of the DNA backbone.[8]
Function
The function of UDG is to remove mutations in DNA, more specifically removing uracil.
Structure
These proteins have a 3-layer alpha/beta/alpha structure. The polypeptide topology of UDG is that of a classic alpha/beta protein. The structure consists primarily of a central, four-stranded, all parallel beta sheet surrounded on either side by a total of eight alpha helices and is termed a parallel doubly wound beta sheet.[9]
Mechanism
Uracil-DNA glycosylases are DNA repair enzymes that excise uracil residues from DNA by cleaving the N-glycosydic bond, initiating the base excision repair pathway. Uracil in DNA can arise either through the deamination of cytosine to form mutagenic U:G mispairs, or through the incorporation of dUMP by DNA polymerase to form U:A pairs.[18] These aberrant uracil residues are genotoxic.[19]
Localisation
In eukaryotic cells, UNG activity is found in both the nucleus and the mitochondria. Human UNG1 protein is transported to both the mitochondria and the nucleus.[20]
Conservation
The sequence of uracil-DNA glycosylase is extremely well conserved[21] in bacteria and eukaryotes as well as in herpes viruses. More distantly related uracil-DNA glycosylases are also found in poxviruses.[22] The N-terminal 77 amino acids of UNG1 seem to be required for mitochondrial localization, but the presence of a mitochondrial transit peptide has not been directly demonstrated. The most N-terminal conserved region contains an aspartic acid residue which has been proposed, based on X-ray structures[23] to act as a general base in the catalytic mechanism.
Family
There are two UDG families, named Family 1 and Family 2. Family 1 is active against uracil in ssDNA and dsDNA. Family 2 excise uracil from mismatches with guanine.[8]
Glycosylases of oxidized bases
A variety of glycosylases have evolved to recognize oxidized bases, which are commonly formed by reactive oxygen species generated during cellular metabolism. The most abundant lesions formed at guanine residues are 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and 8-oxoguanine. Due to mispairing with adenine during replication, 8-oxoG is highly mutagenic, resulting in G to T transversions. Repair of this lesion is initiated by the bifunctional DNA glycosylase OGG1, which recognizes 8-oxoG paired with C. hOGG1 is a bifunctional glycosylase that belongs to the helix-hairpin-helix (HhH) family. MYH recognizes adenine mispaired with 8-oxoG but excises the A, leaving the 8-oxoG intact. OGG1 knockout mice do not show an increased tumor incidence, but accumulate 8-oxoG in the liver as they age.[24] A similar phenotype is observed with the inactivation of MYH, but simultaneous inactivation of both MYH and OGG1 causes 8-oxoG accumulation in multiple tissues including lung and small intestine.[25] In humans, mutations in MYH are associated with increased risk of developing colon polyps and colon cancer. In addition to OGG1 and MYH, human cells contain three additional DNA glycosylases, NEIL1, NEIL2, and NEIL3. These are homologous to bacterial Nei, and their presence likely explains the mild phenotypes of the OGG1 and MYH knockout mice.
Glycosylases of alkylated bases
This group includes E. coli AlkA and related proteins in higher eukaryotes. These glycosylases are monofunctional and recognize methylated bases, such as 3-methyladenine.
AlkA
AlkA refers to 3-methyladenine DNA glycosylase II.[26]
Pathology
- DNA glycosylases involved in base excision repair (BER) may be associated with cancer risk in BRCA1 and BRCA2 mutation carriers.[27]
Epigenetic deficiencies in cancers
Epigenetic alterations (epimutations) in DNA glycosylase genes have only recently begun to be evaluated in a few cancers, compared to the numerous previous studies of epimutations in genes acting in other DNA repair pathways (such as MLH1 in mismatch repair and MGMT in direct reversal).[citation needed] Two examples of epimutations in DNA glycosylase genes that occur in cancers are summarized below.
MBD4
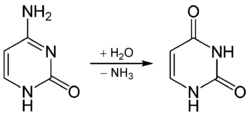
MBD4 (methyl-CpG-binding domain protein 4) is a glycosylase employed in an initial step of base excision repair. MBD4 protein binds preferentially to fully methylated CpG sites.[28] These altered bases arise from the frequent hydrolysis of cytosine to uracil (see image) and hydrolysis of 5-methylcytosine to thymine, producing G:U and G:T base pairs.[29] If the improper uracils or thymines in these base pairs are not removed before DNA replication, they will cause transition mutations. MBD4 specifically catalyzes the removal of T and U paired with guanine (G) within CpG sites.[30] This is an important repair function since about 1/3 of all intragenic single base pair mutations in human cancers occur in CpG dinucleotides and are the result of G:C to A:T transitions.[30][31] These transitions comprise the most frequent mutations in human cancer. For example, nearly 50% of somatic mutations of the tumor suppressor gene p53 in colorectal cancer are G:C to A:T transitions within CpG sites.[30] Thus, a decrease in expression of MBD4 could cause an increase in carcinogenic mutations.
MBD4 expression is reduced in almost all colorectal neoplasms due to methylation of the promoter region of MBD4.[32] Also MBD4 is deficient due to mutation in about 4% of colorectal cancers,[33]
A majority of histologically normal fields surrounding neoplastic growths (adenomas and colon cancers) in the colon also show reduced MBD4 mRNA expression (a field defect) compared to histologically normal tissue from individuals who never had a colonic neoplasm.[32] This finding suggests that epigenetic silencing of MBD4 is an early step in colorectal carcinogenesis.
In a Chinese population that was evaluated, the MBD4 Glu346Lys polymorphism was associated with about a 50% reduced risk of cervical cancer, suggesting that alterations in MBD4 is important in this cancer.[34]
NEIL1
Nei-like (NEIL) 1 is a DNA glycosylase of the Nei family (which also contains NEIL2 and NEIL3).[35] NEIL1 is a component of the DNA replication complex needed for surveillance of oxidized bases before replication, and appears to act as a “cowcatcher” to slow replication until NEIL1 can act as a glycosylase and remove the oxidatively damaged base.[35]
NEIL1 protein recognizes (targets) and removes certain oxidatively-damaged bases and then incises the abasic site via β,δ elimination, leaving 3′ and 5′ phosphate ends. NEIL1 recognizes oxidized pyrimidines, formamidopyrimidines, thymine residues oxidized at the methyl group, and both stereoisomers of thymine glycol.[36] The best substrates for human NEIL1 appear to be the hydantoin lesions, guanidinohydantoin, and spiroiminodihydantoin that are further oxidation products of 8-oxoG. NEIL1 is also capable of removing lesions from single-stranded DNA as well as from bubble and forked DNA structures. A deficiency in NEIL1 causes increased mutagenesis at the site of an 8-oxo-Gua:C pair, with most mutations being G:C to T:A transversions.[37]
A study in 2004 found that 46% of primary gastric cancers had reduced expression of NEIL1 mRNA, though the mechanism of reduction was not known.[38] This study also found that 4% of gastric cancers had mutations in the NEIL1 gene. The authors suggested that low NEIL1 activity arising from reduced expression and/or mutation of the NEIL1 gene was often involved in gastric carcinogenesis.
A screen of 145 DNA repair genes for aberrant promoter methylation was performed on head and neck squamous cell carcinoma (HNSCC) tissues from 20 patients and from head and neck mucosa samples from 5 non-cancer patients.[39] This screen showed that the NEIL1 gene had substantially increased hypermethylation, and of the 145 DNA repair genes evaluated, NEIL1 had the most significantly different frequency of methylation. Furthermore, the hypermethylation corresponded to a decrease in NEIL1 mRNA expression. Further work with 135 tumor and 38 normal tissues also showed that 71% of HNSCC tissue samples had elevated NEIL1 promoter methylation.[39]
When 8 DNA repair genes were evaluated in non-small cell lung cancer (NSCLC) tumors, 42% were hypermethylated in the NEIL1 promoter region.[40] This was the most frequent DNA repair abnormality found among the 8 DNA repair genes tested. NEIL1 was also one of six DNA repair genes found to be hypermethylated in their promoter regions in colorectal cancer.[41]
References
- ↑ Lindahl, T. (1986). "DNA Glycosylases in DNA Repair". Mechanisms of DNA Damage and Repair. 38. 335–340. doi:10.1007/978-1-4615-9462-8_36. ISBN 978-1-4615-9464-2.
- ↑ Aguis, F.; Kapoor, A; Zhu, J-K (2006). "Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation". Proc. Natl. Acad. Sci. U.S.A. 103 (31): 11796–11801. doi:10.1073/pnas.0603563103. PMID 16864782. Bibcode: 2006PNAS..10311796A.
- ↑ Choi, C-S.; Sano, H. (2007). "Identification of tobacco genes encoding proteins possessing removal activity of 5-methylcytosines from intact tobacco DNA". Plant Biotechnology 24 (3): 339–344. doi:10.5511/plantbiotechnology.24.339.
- ↑ 4.0 4.1 "DNA glycosylase recognition and catalysis". Current Opinion in Structural Biology 14 (1): 43–9. February 2004. doi:10.1016/j.sbi.2004.01.003. PMID 15102448.
- ↑ "Atomic structure of the DNA repair [4Fe-4S] enzyme endonuclease III". Science 258 (5081): 434–40. October 1992. doi:10.1126/science.1411536. PMID 1411536. Bibcode: 1992Sci...258..434K.
- ↑ "Human DNA glycosylases involved in the repair of oxidatively damaged DNA". Biol. Pharm. Bull. 27 (4): 480–5. April 2004. doi:10.1248/bpb.27.480. PMID 15056851.
- ↑ "Biochemical characterization and DNA repair pathway interactions of Mag1-mediated base excision repair in Schizosaccharomyces pombe". Nucleic Acids Res. 33 (3): 1123–31. 2005. doi:10.1093/nar/gki259. PMID 15722486.
- ↑ 8.0 8.1 8.2 Pearl LH (2000). "Structure and function in the uracil-DNA glycosylase superfamily.". Mutat Res 460 (3–4): 165–81. doi:10.1016/S0921-8777(00)00025-2. PMID 10946227.
- ↑ 9.0 9.1 "Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis". Cell 80 (6): 869–78. March 1995. doi:10.1016/0092-8674(95)90290-2. PMID 7697717.
- ↑ "Thermostable uracil-DNA glycosylase from Thermotoga maritima a member of a novel class of DNA repair enzymes". Curr. Biol. 9 (10): 531–4. May 1999. doi:10.1016/S0960-9822(99)80237-1. PMID 10339434.
- ↑ "Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions". Cell 92 (1): 117–29. January 1998. doi:10.1016/S0092-8674(00)80904-6. PMID 9489705.
- ↑ "The cap-binding protein complex in uninfected and poliovirus-infected HeLa cells". J. Biol. Chem. 262 (28): 13599–606. October 1987. doi:10.1016/S0021-9258(19)76470-9. PMID 2820976.
- ↑ "Mutational analysis of the damage-recognition and catalytic mechanism of human SMUG1 DNA glycosylase". Nucleic Acids Res 32 (17): 5291–5302. 2004. doi:10.1093/nar/gkh859. PMID 15466595.
- ↑ Wu, Peiying; Qiu, Chen; Sohail, Anjum; Zhang, Xing; Bhagwat, Ashok S.; Cheng, Xiaodong (2003-02-14). "Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4". The Journal of Biological Chemistry 278 (7): 5285–5291. doi:10.1074/jbc.M210884200. ISSN 0021-9258. PMID 12456671.
- ↑ Wong E et al. (1995). "Mbd4 inactivation increases C→T transition mutations and promotes gastrointestinal tumor formation". PNAS 99 (23): 14937–14942. doi:10.1073/pnas.232579299. PMID 12417741.
- ↑ "Crystal structure and mutational analysis of human uracil-DNA glycosylase". Cell 80 (6): 869–878. 1995. doi:10.1016/0092-8674(95)90290-2. PMID 7697717.
- ↑ Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. (1996). A nucleotide-flipping mechanism from the structure of human uracil–DNA glycosylase bound to DNA. 384: 87-92.
- ↑ "Uracil in DNA--general mutagen, but normal intermediate in acquired immunity". DNA Repair (Amst.) 6 (4): 505–16. April 2007. doi:10.1016/j.dnarep.2006.10.014. PMID 17116429.
- ↑ Hagen L; Peña-Diaz J; Kavli B; Otterlei M; Slupphaug G; Krokan HE (August 2006). "Genomic uracil and human disease". Exp. Cell Res. 312 (14): 2666–72. doi:10.1016/j.yexcr.2006.06.015. PMID 16860315.
- ↑ "Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene". Nucleic Acids Res. 21 (11): 2579–84. June 1993. doi:10.1093/nar/21.11.2579. PMID 8332455.
- ↑ "Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme". EMBO J. 8 (10): 3121–5. October 1989. doi:10.1002/j.1460-2075.1989.tb08464.x. PMID 2555154.
- ↑ "Identification of a poxvirus gene encoding a uracil DNA glycosylase". Proc. Natl. Acad. Sci. U.S.A. 90 (10): 4518–22. May 1993. doi:10.1073/pnas.90.10.4518. PMID 8389453. Bibcode: 1993PNAS...90.4518U.
- ↑ "The structural basis of specific base-excision repair by uracil-DNA glycosylase". Nature 373 (6514): 487–93. February 1995. doi:10.1038/373487a0. PMID 7845459. Bibcode: 1995Natur.373..487S.
- ↑ Klungland A et al. (1999). "Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage". PNAS 96 (23): 13300–13305. doi:10.1073/pnas.96.23.13300. PMID 10557315. Bibcode: 1999PNAS...9613300K.
- ↑ Russo, Maria Teresa et al. (2004). "Accumulation of the Oxidative Base Lesion 8-Hydroxyguanine in DNA of Tumor-Prone Mice Defective in Both the Myh and Ogg1 DNA Glycosylases". Cancer Res 64 (13): 4411–4414. doi:10.1158/0008-5472.can-04-0355. PMID 15231648.
- ↑ "Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans". Acta Crystallogr D 68 (6): 703–12. 2012. doi:10.1107/S090744491200947X. PMID 22683793.
- ↑ Osorio, A; Milne, R. L.; Kuchenbaecker, K; Vaclová, T; Pita, G; Alonso, R; Peterlongo, P; Blanco, I et al. (2014). "DNA Glycosylases Involved in Base Excision Repair May Be Associated with Cancer Risk in BRCA1 and BRCA2 Mutation Carriers". PLOS Genetics 10 (4): e1004256. doi:10.1371/journal.pgen.1004256. PMID 24698998.
- ↑ Walavalkar, Ninad (2014). "Solution structure and intramolecular exchange of methyl-cytosine binding domain protein 4 (MBD4) on DNA suggests a mechanism to scan for mCpG/TpG mismatches". Nucleic Acids Research 42 (17): 11218–11232. doi:10.1093/nar/gku782. PMID 25183517.
- ↑ "Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites". DNA Repair 32: 33–42. Aug 2015. doi:10.1016/j.dnarep.2015.04.011. PMID 26021671.
- ↑ 30.0 30.1 30.2 "MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles". Mutation Research 743–744: 12–25. 2013. doi:10.1016/j.mrfmmm.2012.11.001. PMID 23195996.
- ↑ "The CpG dinucleotide and human genetic disease". Human Genetics 78 (2): 151–5. Feb 1988. doi:10.1007/bf00278187. PMID 3338800.
- ↑ 32.0 32.1 "Epigenetic downregulation of the DNA repair gene MED1/MBD4 in colorectal and ovarian cancer". Cancer Biology & Therapy 8 (1): 94–100. Jan 2009. doi:10.4161/cbt.8.1.7469. PMID 19127118.
- ↑ "Involvement of MBD4 inactivation in mismatch repair-deficient tumorigenesis". Oncotarget 6 (40): 42892–904. Oct 2015. doi:10.18632/oncotarget.5740. PMID 26503472. PMC 4767479. https://dash.harvard.edu/bitstream/handle/1/26318553/4767479.pdf?sequence=1.
- ↑ "The MBD4 Glu346Lys polymorphism is associated with the risk of cervical cancer in a Chinese population". Int. J. Gynecol. Cancer 22 (9): 1552–6. 2012. doi:10.1097/IGC.0b013e31826e22e4. PMID 23027038.
- ↑ 35.0 35.1 "Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins". Proc. Natl. Acad. Sci. U.S.A. 110 (33): E3090–9. 2013. doi:10.1073/pnas.1304231110. PMID 23898192. Bibcode: 2013PNAS..110E3090H.
- ↑ "Variant base excision repair proteins: contributors to genomic instability". Seminars in Cancer Biology 20 (5): 320–8. Oct 2010. doi:10.1016/j.semcancer.2010.10.010. PMID 20955798.
- ↑ "Effects of base excision repair proteins on mutagenesis by 8-oxo-7,8-dihydroguanine (8-hydroxyguanine) paired with cytosine and adenine". DNA Repair (Amst.) 9 (5): 542–50. 2010. doi:10.1016/j.dnarep.2010.02.004. PMID 20197241.
- ↑ "Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer". Carcinogenesis 25 (12): 2311–7. 2004. doi:10.1093/carcin/bgh267. PMID 15319300.
- ↑ 39.0 39.1 "Epigenetic screen of human DNA repair genes identifies aberrant promoter methylation of NEIL1 in head and neck squamous cell carcinoma". Oncogene 31 (49): 5108–16. 2012. doi:10.1038/onc.2011.660. PMID 22286769.
- ↑ "A critical re-assessment of DNA repair gene promoter methylation in non-small cell lung carcinoma". Scientific Reports 4: 4186. 2014. doi:10.1038/srep04186. PMID 24569633. Bibcode: 2014NatSR...4E4186D.
- ↑ "DNA methylation changes in genes frequently mutated in sporadic colorectal cancer and in the DNA repair and Wnt/β-catenin signaling pathway genes". Epigenomics 6 (2): 179–91. Apr 2014. doi:10.2217/epi.14.7. PMID 24811787.
External links
- DNA+Glycosylases at the US National Library of Medicine Medical Subject Headings (MeSH)
 |